Evolutionary genomics of adaptation and diversification
Lab News:
We’re looking for a Postdoc! Apply here if you’re interested in joining our team!
October 2024: We are looking for two PhD students to join the lab in 2025. Feel free to email if you have any questions!
August 2024: Congratulations to Rachel O’Dell who left us to pursue her PhD with the Ingram-Smith lab!
July 2024: Congratulations to Dr. Brianna Ports who has left us for a tenure-track appointment at Carlow University!
July 2024: We were awarded an NSF CAREER grant for work on wing pattern mimicry evolution in heliconiine butterflies!
See some of our highlighted recent publications below:
Recent publications:
The Dryas iulia Genome Supports Multiple Gains of a W Chromosome from a B Chromosome in Butterflies
Assembled sex chromosomes are important for studies on the evolution of sex, sex-linked traits, and sex-biased gene expression. The Dryas iulia genome provides an important case study on the origin and evolution of ZZ/ZW sex determination systems from a ZZ/Z0 ancestral state. Our findings support a B chromosome origin for the W chromosome and provide evidence that W chromosomes likely originated from B chromosomes multiple times in butterflies. These results indicate that sex determination mechanisms and sex chromosomes may turnover rapidly when sex chromosome pairs have nonhomologous origins. Genome Biology and Evolution (2021) 13 (7) evab128.
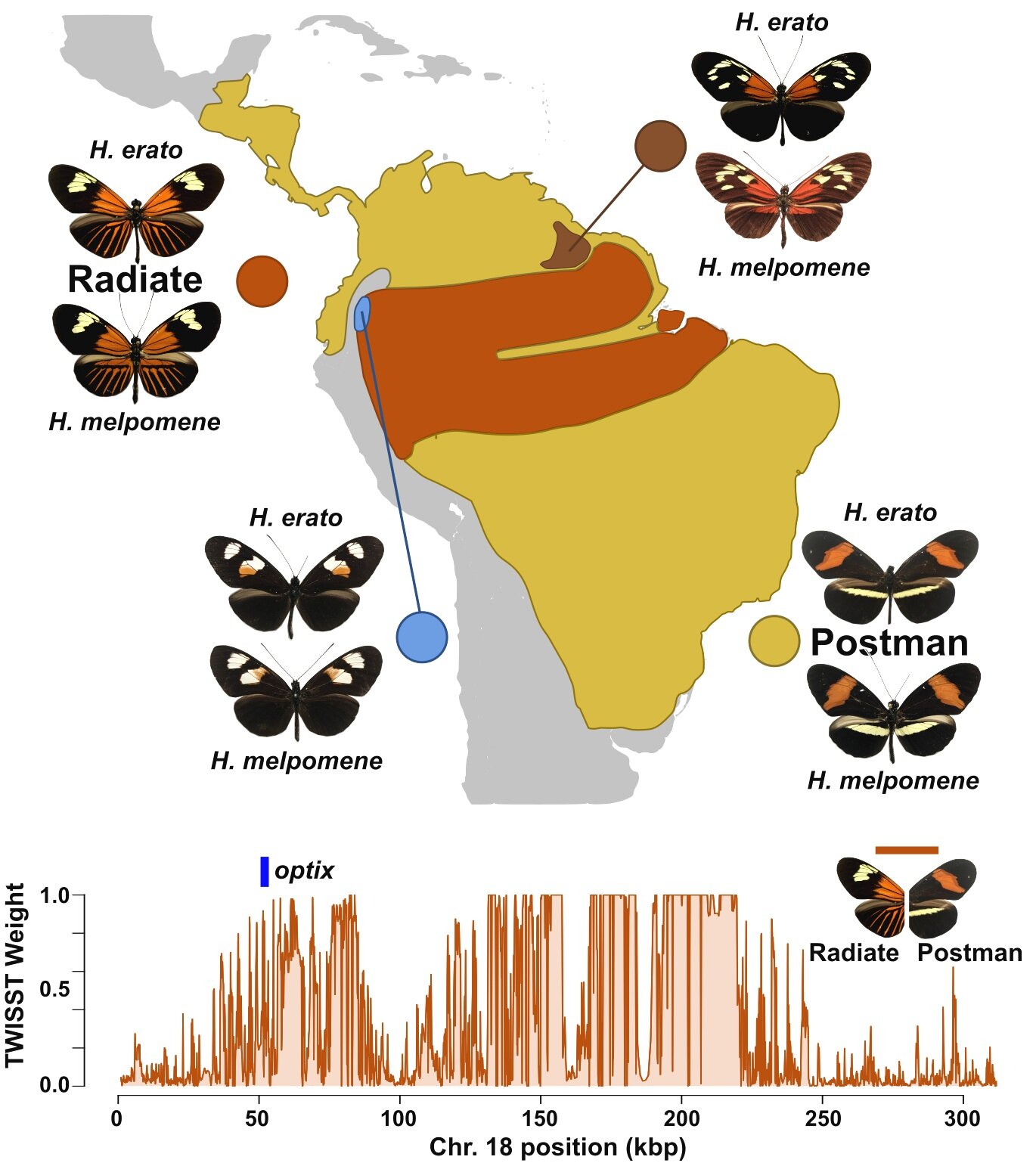
Many functionally connected loci foster adaptive diversification along a neotropical hybrid zone.
Recent studies of adaptation frequently propose a few large-effect loci for the genetic basis of novel phenotypes. Yet these results are often at odds with quantitative studies finding a complex, multigenic architecture for many traits. To determine whether adaptation of a Mendelian phenotype may still be a multigenic process, we studied the red color pattern gene network in Heliconius butterflies. We find that the red wing pattern master regulator gene Optix binds dozens of loci under selection, and these loci collectively maintain separation between adaptive red color pattern phenotypes in natural populations. We propose a model of trait evolution where functional connections between loci may resolve much of the disparity between large-effect and multigenic evolutionary models. Science Advances (2020) 6 (39) eabb8617.
Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry.
Color pattern mimicry in Heliconius butterflies provides a key example of trait evolution via selection on large effect genes. To understand how individual genes drive the evolution of new, complex traits, we functionally characterized 5 enhancers of the red color pattern gene in Heliconius. We discovered these enhancers are all necessary for color patterning and influence multiple elements of wing patterning rather than driving distinct wing pattern modules. Enhancer variants are shared among similarly colored disjunct populations via hybridization. These enhancers also appear to be evolving in parallel in distantly related co-mimic species, suggesting their ancient role in Heliconius wing pattern diversity. We propose a model for how interacting enhancers and their upstream modifiers can generate wing pattern diversity. PNAS (2019) 116 (48) 24174-24183.